Chemical Science & Engineering Research
Title
Development of Interpretable QSAR Model for Quick Screening of Inhibitors against Tyrosine Protein Kinase JAK-2
Authors
Sharav Desai*a and Dhananjay Meshramb
aDepartment of Pharmaceutical Microbiology and Biotechnology, Pioneer Pharmacy Degree College, Vadodara-390019, Gujarat, India.
bDepartment of Quality assurance, Pioneer Pharmacy Degree College, Vadodara-390019, Gujarat, India.
*Corresponding author E-mail address: Sharavdesai@gmail.com (Sharav Desai)
Article History
Publication details: Received: 01st May 2022; Revised: 15th June 2022; Accepted: 15th June 2022; Published: 04th July 2022
Cite this article
Desai S.; Meshram D. Development of Interpretable QSAR Model for Quick Screening of Inhibitors against Tyrosine Protein Kinase JAK-2. Chem. Sci. Eng. Res., 2022, 4(10), 46-53.
Abstract
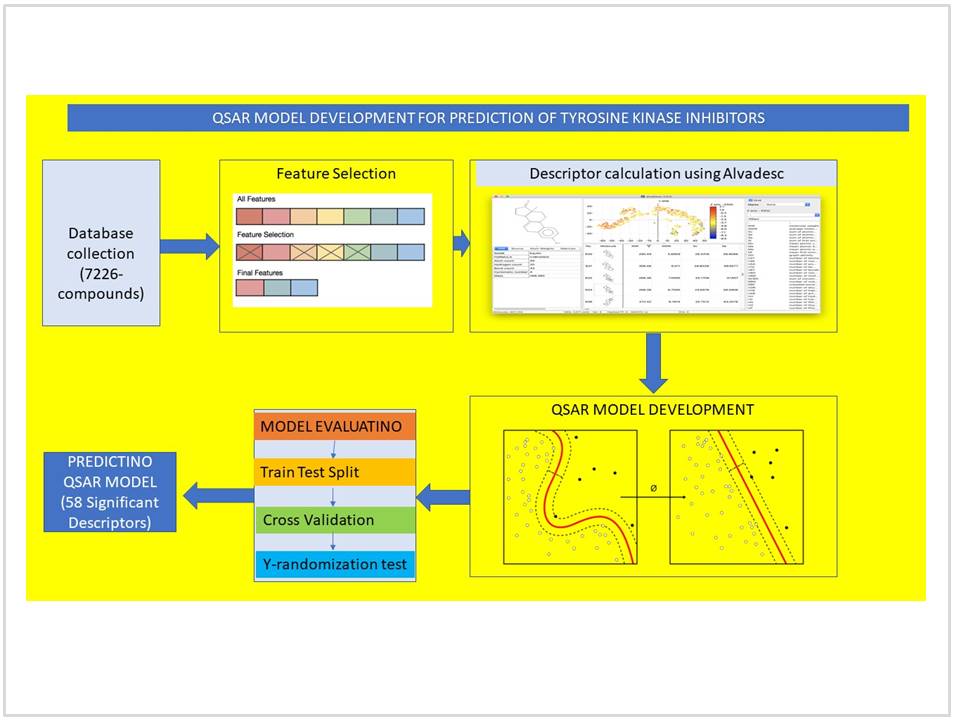
Kinase belongs to large family of enzymes that catalyse transfer of high energy phosphate molecule to substrates like protein, lipids, carbohydrates and nucleic acid. Protein tyrosine kinases are becoming therapeutically active target as It plays a significant role in several signal transduction and immunological reactions. Dysregulation, overexpression and mutation of protein kinase found in many diseases including cancer and immunopathological conditions. InSilico methods of drug discovery are considerably cheaper and faster compared to traditional methods available today. In the present work, the use of QSAR model is shown in the discovery of new tyrosine kinase inhibitors. Total of 7226 compounds retrieved from the ChEMBL database and were used after manual curation. More than 2000 descriptors of different class were calculated for individual compounds. Manual curation, outlier removal and feature selection techniques were used to reduce the number of insignificant features. Four machine learning algorithms called SVR, MLR, RF and RT are used to build the final QSAR model. We also have applied the internal and external evaluation parameters to check the model stability and its prediction power. All the four models developed were showing acceptable range of R2 like 59.40, 58.84, 97.1, and 99.32 for MLR, SVR, RF and RT respectively on training set. Similarly test dataset was evaluated with the same matrix and showing nearly similar values to train set except RT algorithm. Y-randomization test also performed and confirmed that model is not produced by chance.
Keywords
Protein tyrosine kinase; QSAR; Cancer; Machine learning; MLR; SVR; RF; RT